TLDR
Three and a half years at OpenEye Scientific (now Cadence Molecular Sciences). Designed 3 interfaces, shipped 1.5. Productized Molecule Search: grew user base 10% (~250→275+) with accessible licensing. Established research practice. Built design system. Shifted shipping velocity 2→7+ releases/year with Principal Product Owner Brent Jenkins.
Challenge: Bridge wet lab (bench chemistry, 2D) and dry lab (computational modeling, 3D). Give medicinal chemists control in unfamiliar environments. Support comprehension at billion-molecule scale. Support creativity of experimentation and clarity of communication throughout project lifecycles.
The Drug Discovery Problem
Drug discovery cycles through distinct phases—target exploration, hit finding, hit-to-lead, lead optimization—each with different computational and experimental needs. Wet lab (bench chemistry, 2D thinking) and dry lab (computational modeling, 3D space) must collaborate across the DMTA cycle: Design, Make, Test, Analyze.
Context evaporates between handoffs. One scientist’s “why I chose this molecule” becomes another’s unexplained starting point. Research showed scientists spending 70% of time assembling presentations for teammates instead of doing science. Scientific reasoning was happening around platforms, not within them.
In addition to this industry-wide continuity challenge, we were designing for a scale challenge - our own computational excellence could overwhelm users if we were not careful:
- FastROCS: 1 billion compounds in <30 minutes (previously: 11 CPU-years or 2 days on 2,000 CPU cluster)
- ROCS X: trillions of molecules using AI-enabled methods
- Different scales require different approaches: at trillions, only certain filters are computationally feasible; at billions, different refinement strategies work; at millions, detailed examination becomes possible
The design tension: Control ↔ Abstraction
How do you give medicinal chemists control in unfamiliar 3D computational environments while preserving power for computational experts? Technical enough for specialists (precision, determinism, tunability). Approachable enough for bench chemists (simplicity, learnability, exploration). Abstraction as conduit for learning, not ceiling on expertise.
In 2022, Cadence (semiconductor design) acquired OpenEye. This presented new opportunities but also new challenges: life sciences are stochastic and exploratory; chip design is deterministic and tightly constrained. About a year later, a large downturn paralyzed the biotech sector. Some called this time period a nuclear winter—frozen funding, zero tolerance for speculative work. Tools had to make uncertainty legible, not just manage it. Our team also had to do more with less. We accomplished a great deal as a very small team, and I am so proud of us for what we accomplished.
Molecule Search: Opening the Platform
My primary contribution: productized Molecule Search—getting the company to open its eyes to what seemed too obvious. Other companies had standalone molecule search capabilities. Our advantage: computational power, algorithmic elegance (ROCS), scientific trust. The team initially overlooked this easy win, preferring harder problems. I pitched and pushed. We shipped in ~1 year.
Before: Orion served computational chemists. Medicinal chemists—who synthesize compounds—couldn’t query billion-molecule databases without intermediaries.
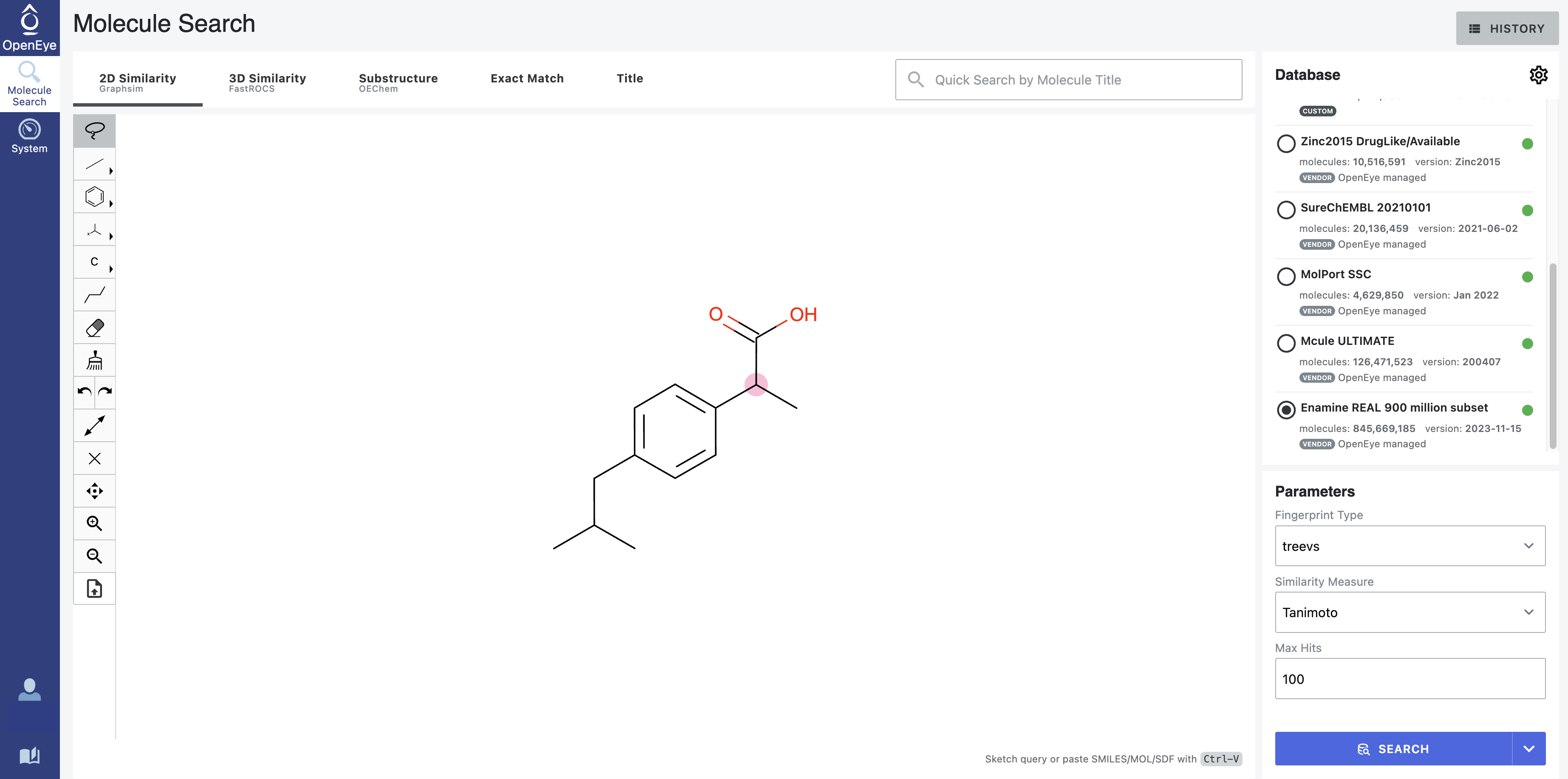
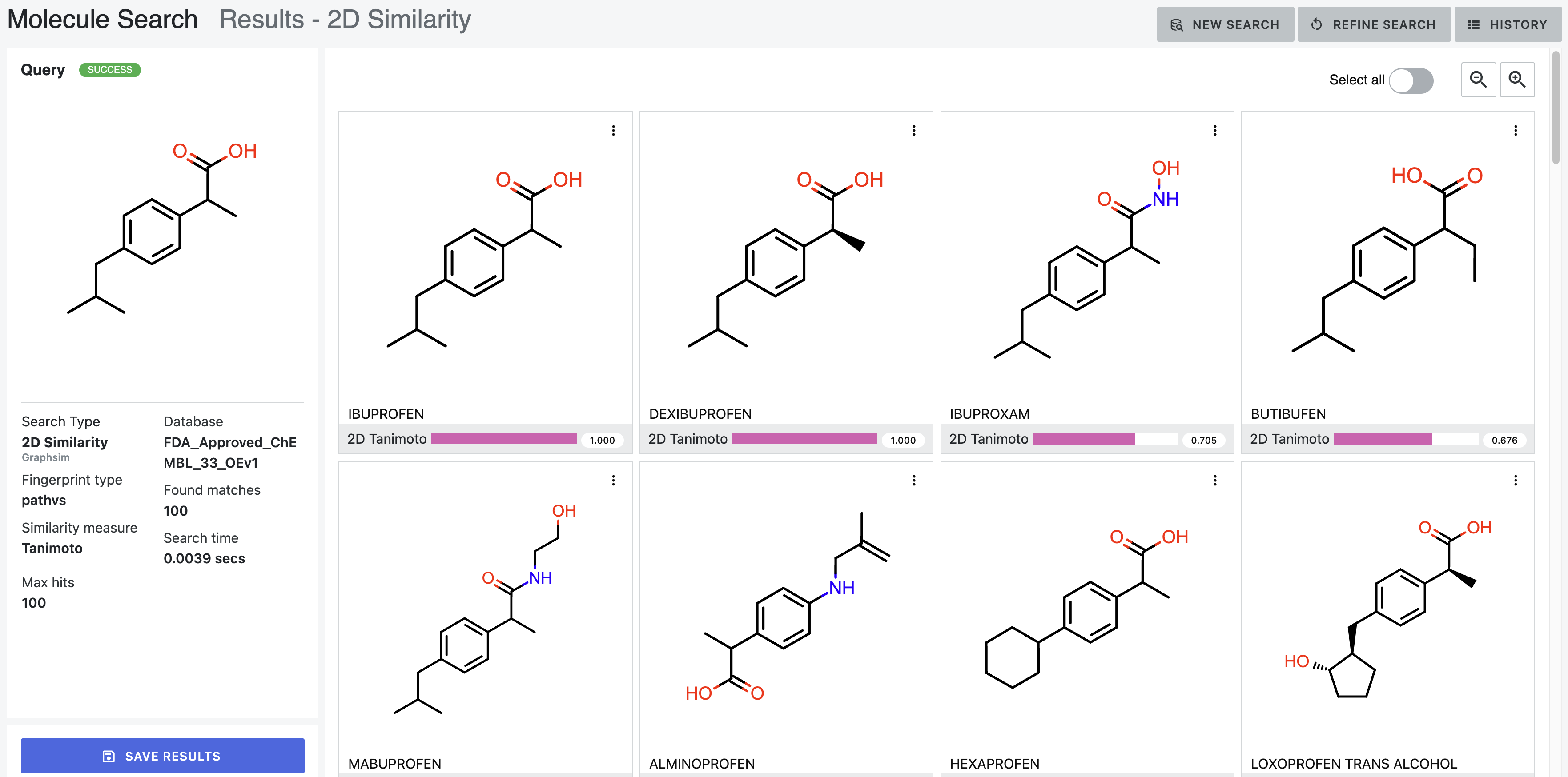
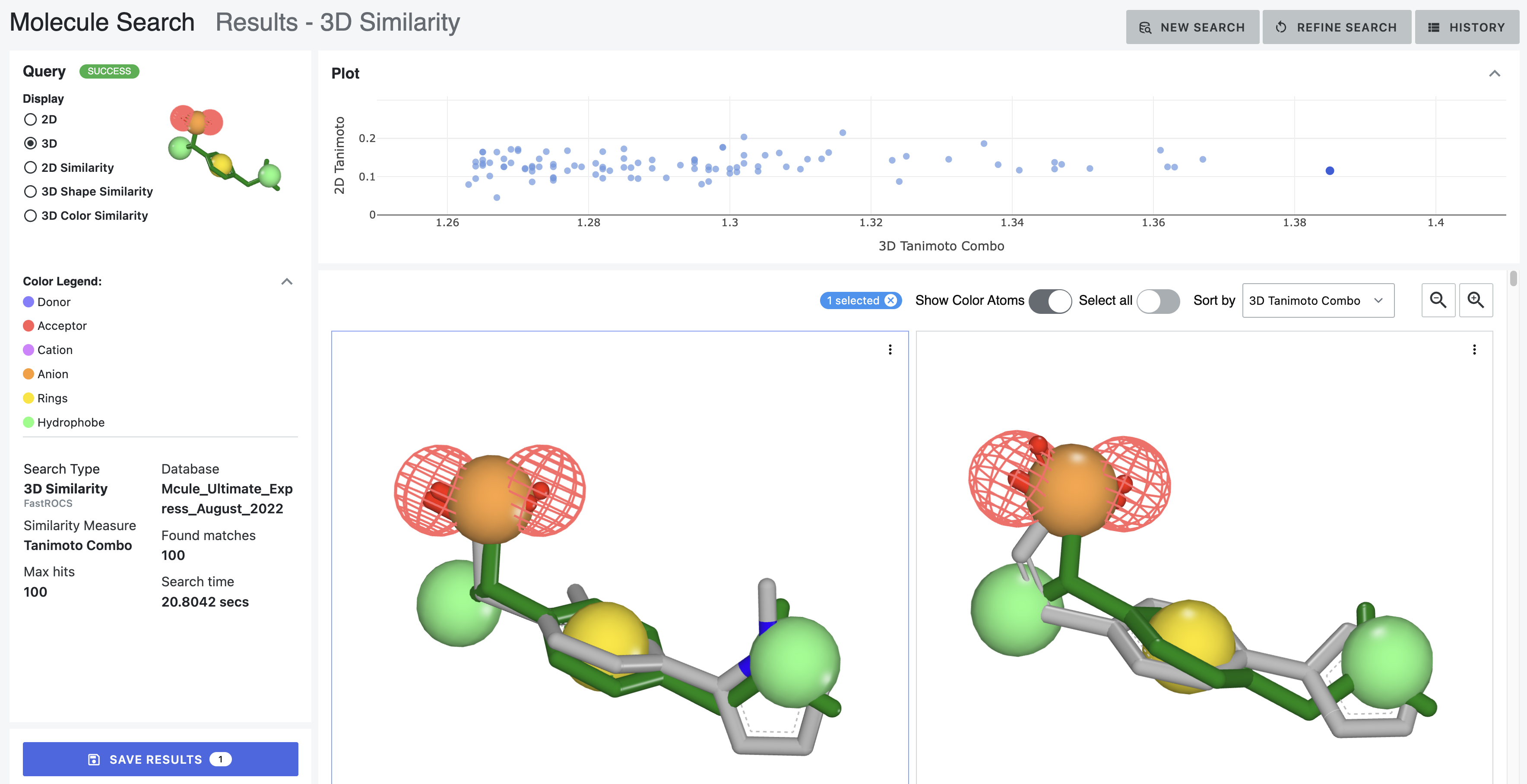
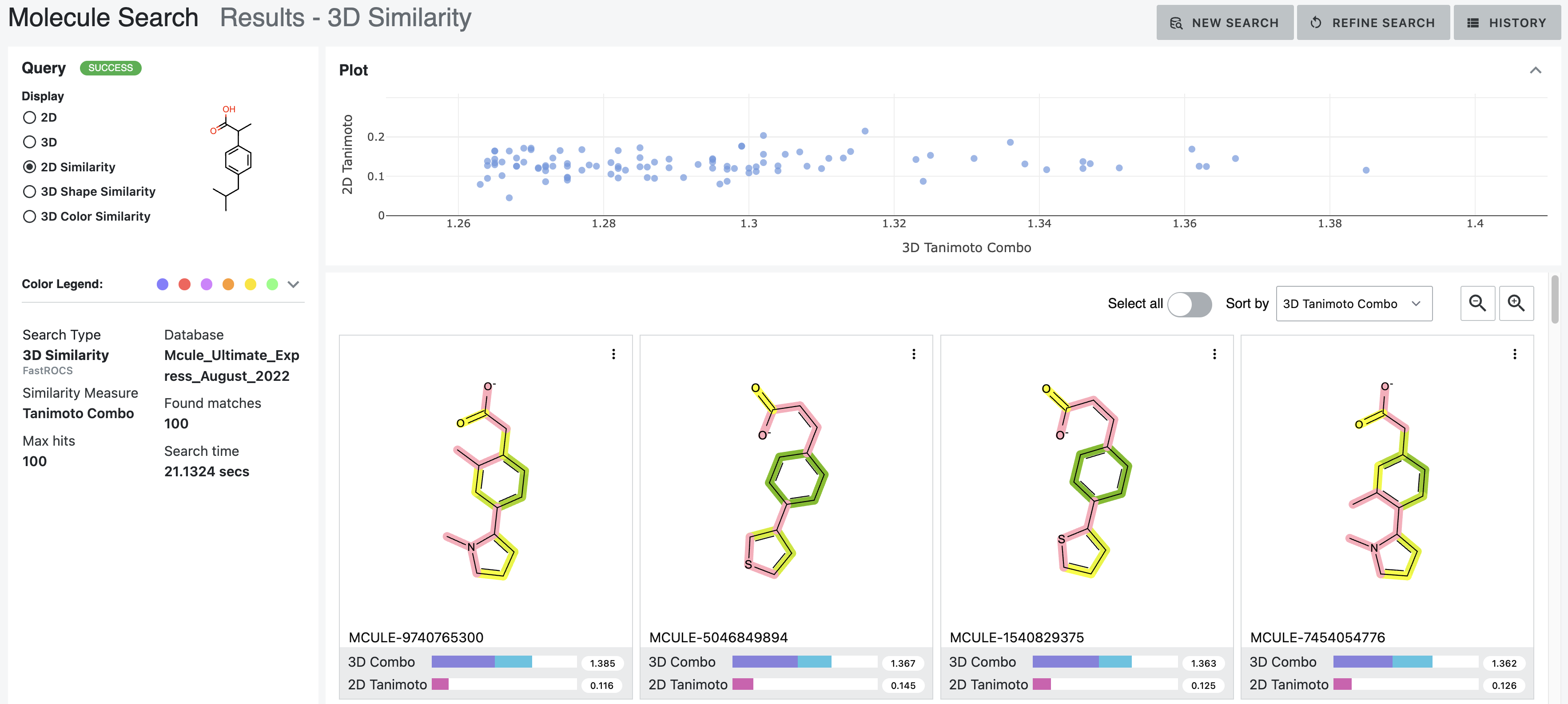
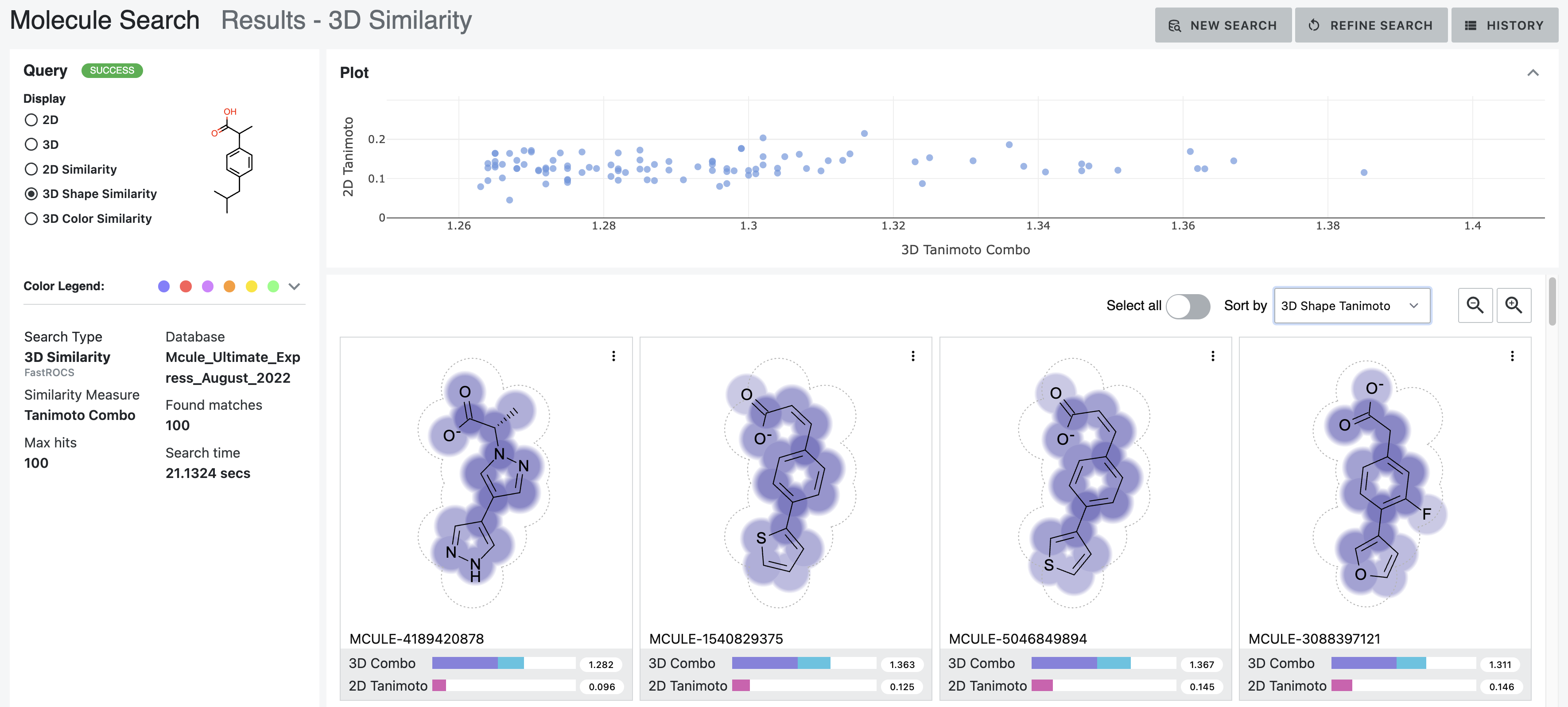
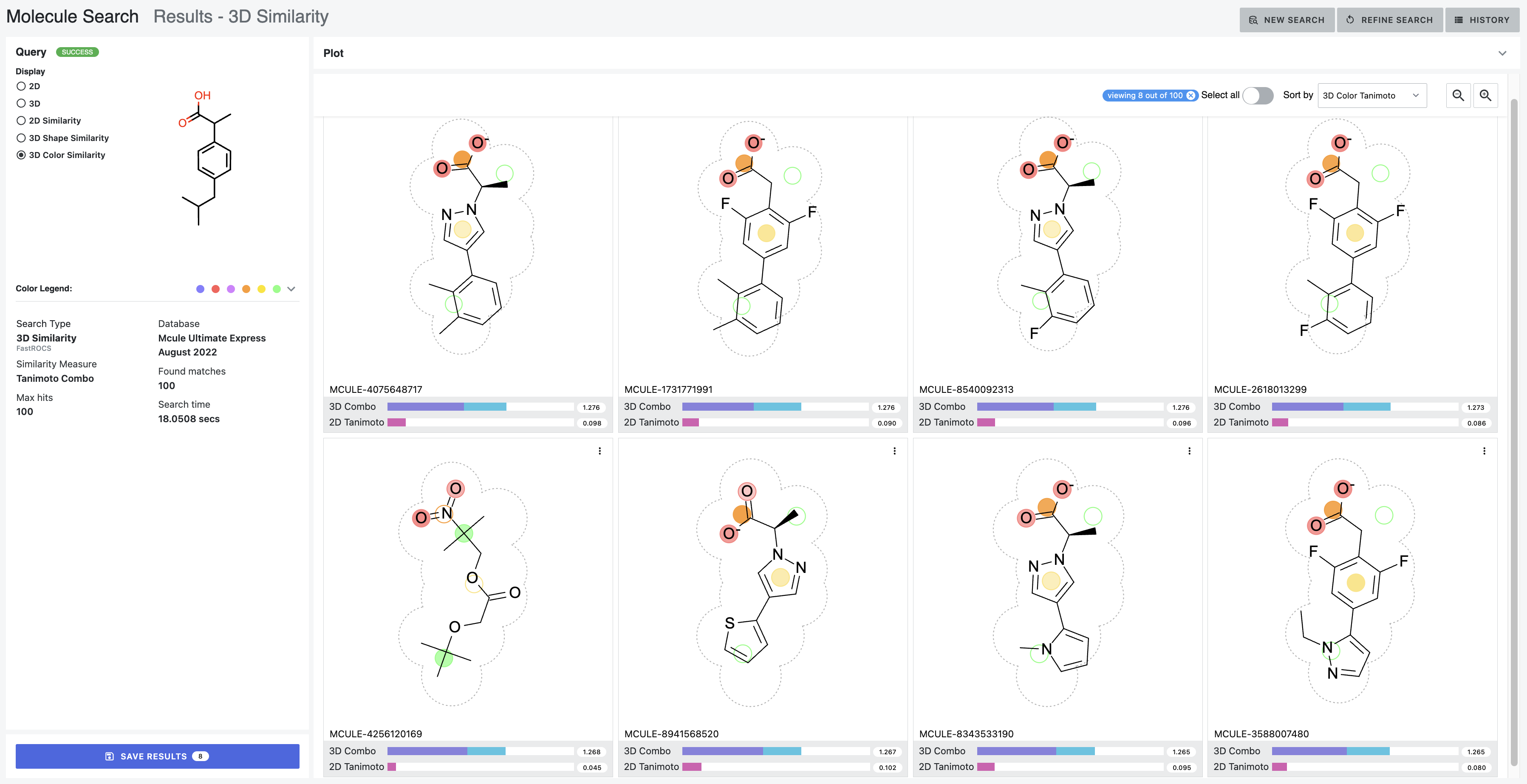
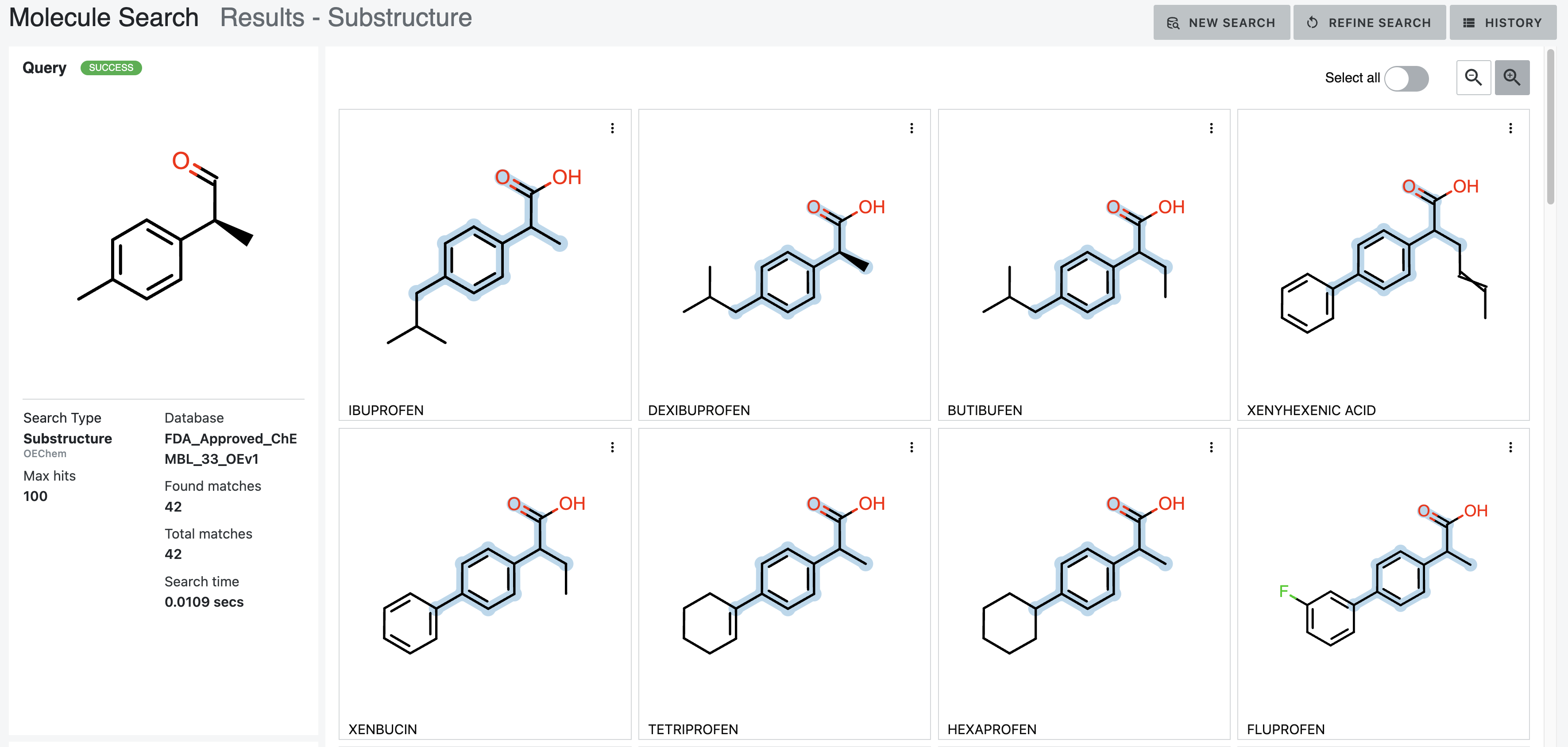
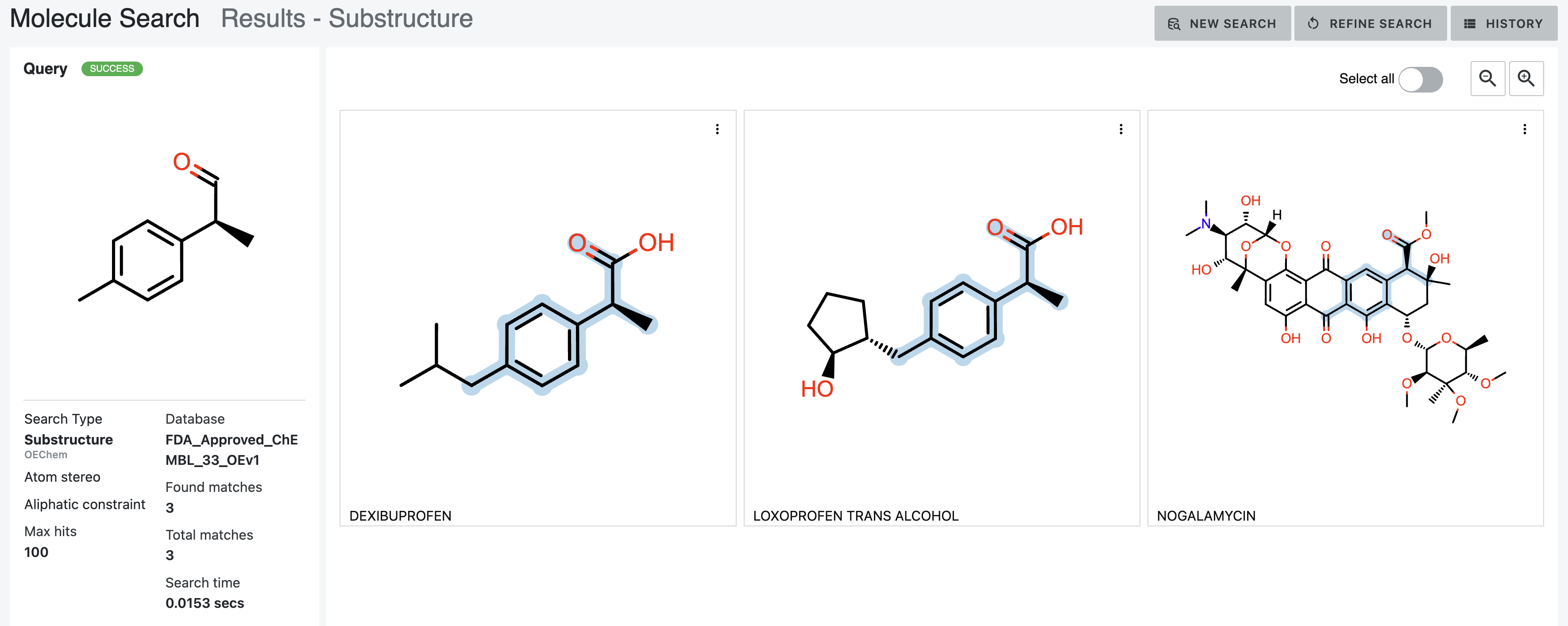
After: Our first tool letting medicinal chemists explore 3D computational chemistry directly. Multi-modal queries (2D similarity, 3D shape, substructure, properties). Search results designed to update in real-time as scientists adjust faceted filter parameters. Medicinal chemists sketch structures; system finds matches at billion-molecule scale. Progressive refinement from broad to specific.
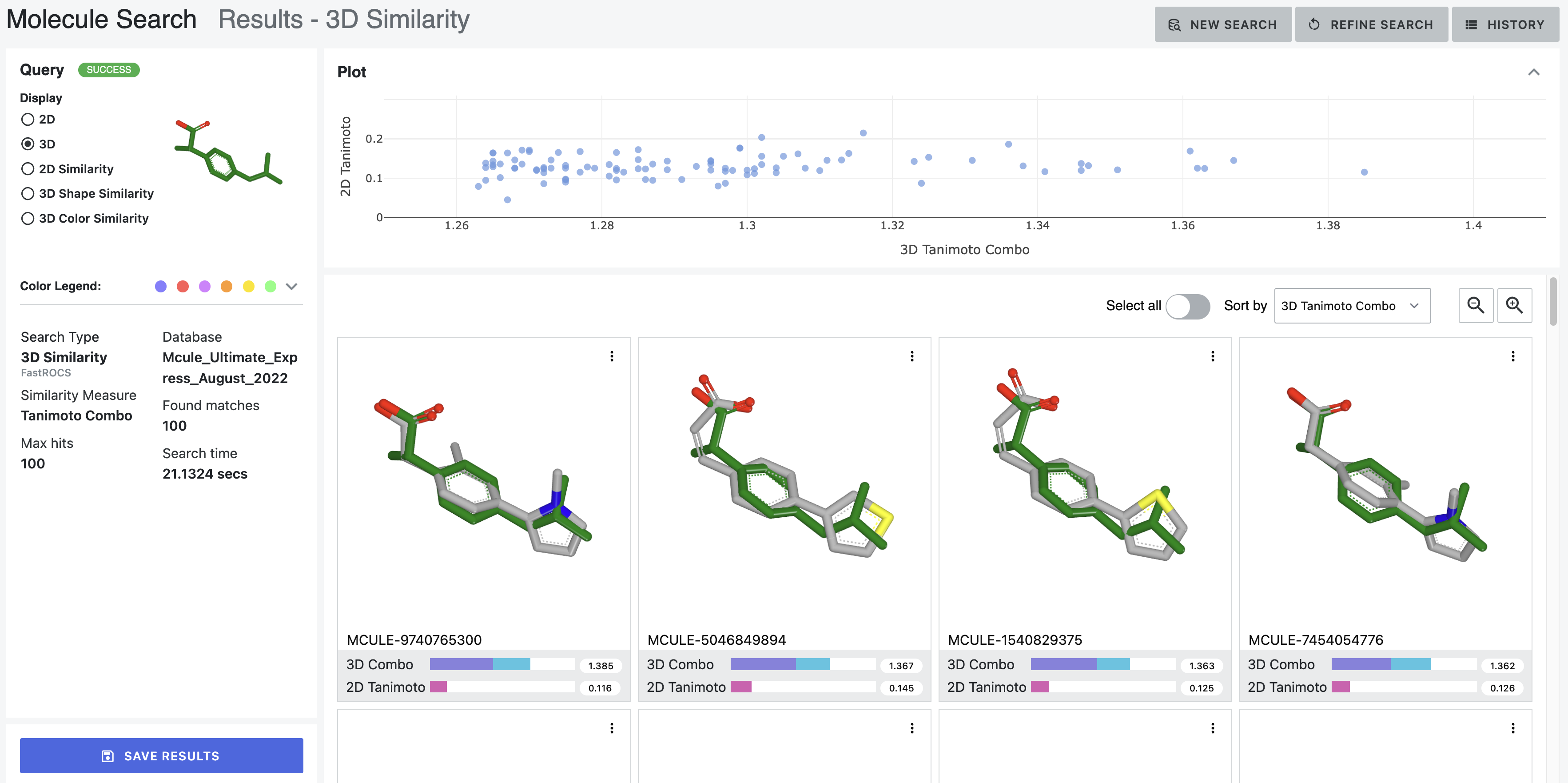
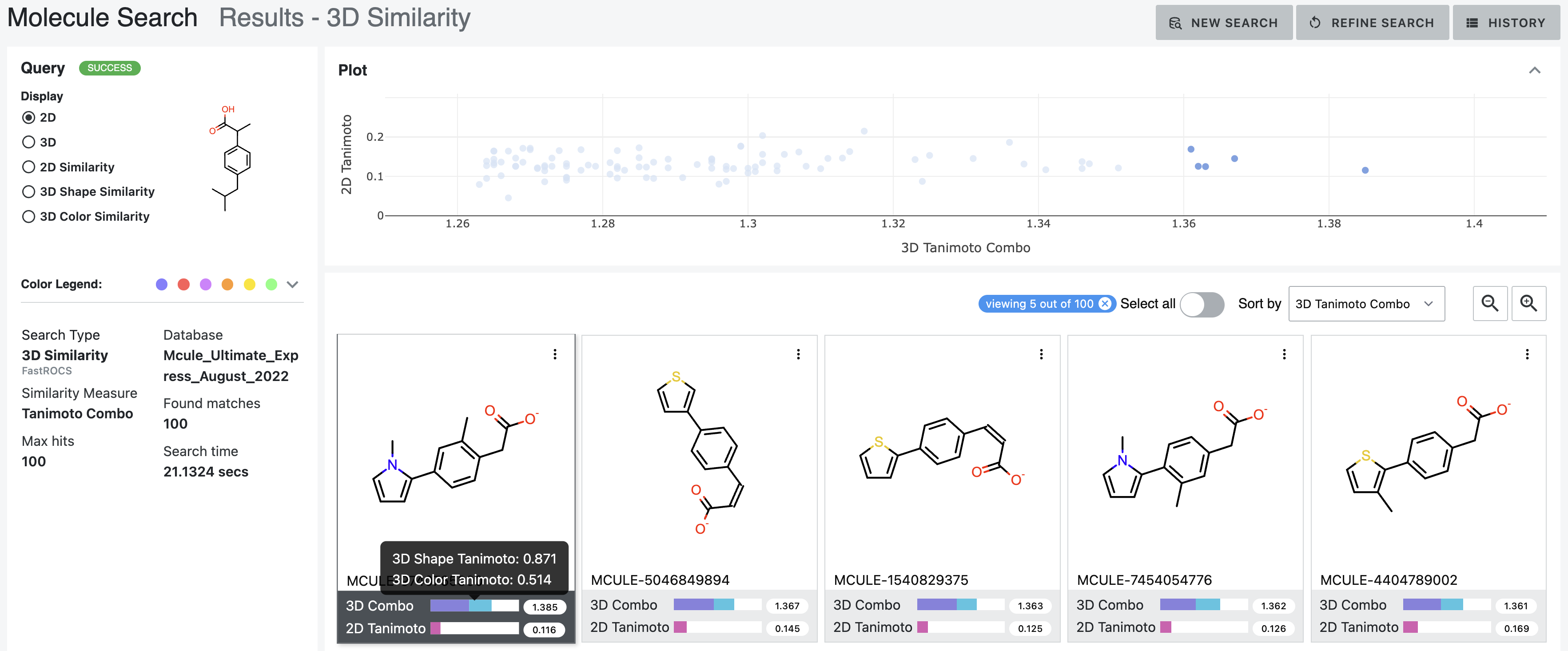
How it works — Multi-modal search combining 2D similarity, 3D shape, substructure patterns, and property filters
Search by 2D structural similarity (molecules sharing chemical scaffolds), 3D shape (geometric similarity, pharmacophore matching), substructure (specific chemical patterns), or property filters (molecular weight, lipophilicity, predicted ADME). Combine all simultaneously.
Sketch what you’re looking for. System finds similar molecules at scale. Refine progressively: “show me everything like this” → “show me only molecules with these exact features and this shape.”
The design challenge — Scale-appropriate refinement across discovery phases, transparent ranking, synthesis integration
Different discovery phases need different tools. Target exploration: broad search. Hit finding: pattern recognition. Hit-to-lead: detailed comparison. Lead optimization: precise property prediction.
Different scales need different refinement. Trillions: certain filters only. Billions: different approaches. Millions: detailed examination. Interface makes ranking transparent—scientists question results, verify logic, override when chemical intuition differs.
Integration with synthesis: finding interesting molecules only matters if you can get them, make them, or buy them. Connected to purchasability, synthetic accessibility, catalog availability.
What changed — 10% user growth, platform repositioning, primary pharma interface
Grew user base 10% (~250→275+) with accessible licensing (thousands vs. much higher full seat cost). Medicinal chemists—largest pharma user population—could interact with computational chemistry directly.
Platform positioning shifted: specialist tool → drug discovery platform. Became primary way pharmaceutical teams interact with Orion for lead identification, scaffold hopping, hit-to-lead optimization.
Platform Work: Control, Creativity, Comprehension
Led platform redesign for interdisciplinary science. Multi-track strategy: exploratory (long-term vision), strategy (wet/dry lab workflows), tactical (incremental improvements), operational (daily velocity). Framework became how organization thought about development.
Designed comprehensive systems; company resources limited what shipped. Most designers ship a fraction of what they design—no apology needed.
Multi-track approach — Four parallel tracks enabling replatforming while shipping continuously
Exploratory: Long-term vision for modeling interfaces, emerging paradigms, uncertainty representation
Strategy: Cross-product workflows, wet lab ↔ dry lab handoffs, DMTA collaboration patterns
Tactical: Feature improvements, workflow optimization, usability fixes compounding over time
Operational: Day-to-day improvements, small wins, maintaining momentum
Kept long-term vision connected to near-term execution.
3D Molecular Modeling — Multi-monitor support, simplified navigation, clarity over complexity
Designed multi-monitor workflows—modeling on one screen, analysis on another, documentation on third.
Simplified complex navigation patterns in online 3D viewer. Prioritized display structures favoring clarity over complexity. Camera controls matching scientific intuition (rotating around binding sites, aligning structures, examining relevant angles), not generic 3D software conventions.
Task-specific representation modes: verification needs detail, exploration needs patterns, presentation needs clarity.
LLM Integration — AI extends judgment, doesn't replace it
Developed principles ensuring AI assistance extended rather than replaced scientific judgment. Prototyped natural language queries, assisted literature search, method recommendations.
Challenge: interpretable enough to trust, transparent enough to verify. Made reasoning visible, questionable. Scientists need to override AI when domain knowledge differs.
Error Messaging — Scientific guidance, not stack traces
Transformed technical failures (stack traces, NaN errors) into scientific guidance. Not “NaN in force calculation”—“Energy minimization couldn’t converge; try adjusting charge model or simplifying starting structure.”
Explained what went wrong scientifically, why it matters, what to try next.
Documentation — Task-oriented, progressively disclosed, education embedded
Research revealed competitor’s docs “made customers feel loved” vs. ours “made people feel like they needed PhD in OpenEye.”
Redesign: task-oriented structure (not just API reference), progressive disclosure (basics → advanced → expert), scientific context (why use this method, not just how), integration examples, education at point of need.
MMDS — Protein preparation as discovery, not barrier
Redesigned how protein structural data gets organized, accessed, prepared for computation. Protein family navigation, structure quality assessment, alignment visualization, docking workflow integration.
Made protein preparation feel like discovery rather than tedious prerequisite.
Research & Velocity
Built research format encouraging inquiry beyond feature requests. As only designer, established practice shifting from purely commercial visits to genuine inquiry about scientific reasoning. Commercial team: greatest supporters—built on their foundation.
Contextual studies with scientists in labs. Domain expert testing. Continuous feedback loops. Trained product managers and engineers to conduct research. Teams now test assumptions before building. Product decisions reference user data. Practice persisted beyond my tenure.
Research questions evolved:
- Not “Does this work?” but “How does this fit into discovery workflow?”
- Not “Is this fast?” but “What are you trying to understand?”
- Not “What do you want?” but “What makes you confident in results?”
Design system: Angular framework (no longer supported) made full rebuild impossible. Created legacy skin strategy—modern visual layer applied progressively to stable architecture.
Result: Working with Brent Jenkins (Principal Product Owner), shifted velocity 2→7+ releases/year. Small improvements compounded. Incremental value beats all-or-nothing bets.
What Changed
Molecule Search: 10% user growth, accessible licensing, direct medicinal chemist access. Platform positioned as drug discovery tool. Became primary pharma interface.
Platform capabilities: ROCS X interface design, continued computational evolution during tenure.
Velocity: 2→7+ releases/year through design system + product leadership collaboration.
Design Principles That Emerged
Control ↔ Abstraction: Technical enough for computational chemists (precision, determinism, tunability). Approachable enough for medicinal chemists (simplicity, learnability). Power revealed progressively, never hidden or dumped. Abstraction as conduit for learning, not ceiling on expertise.
Respect Human Time Scales: Scientific work operates across rhythms—instant (<1s), interactive (<15s), coffee (<5min), lunch, overnight, HPC. Interfaces must acknowledge this. Speed without control creates ambiguity and chaos, sometimes disaster.
Preserve Context Across DMTA: Design-Make-Test-Analyze handoffs lose reasoning. Platforms need to preserve “why this molecule” across wet lab and dry lab collaboration. Scientific reasoning should happen within platforms, not around them.
Scale-Appropriate Refinement: Trillions→billions→millions requires different approaches. Target exploration→hit finding→hit-to-lead→lead optimization needs different tools. FastROCS: 1B compounds <30min vs. 11 CPU-years previously. ROCS X: trillions. Interface supports all scales with appropriate refinement strategies.
Scientists Think Through Models: Computational chemists don’t just use models—they think through them. Interface shapes what’s thinkable. Design challenge: preserve representational fidelity while making tools operable.
Work Within Constraints: Systems reflect their moment’s constraints—technical, cultural, economic. Sometimes the effective approach is working intelligently within limits rather than against them.
What Continues
ACME STEAM Co.: modeling and simulation tools for interdisciplinary science.
Core questions:
- Make modeling accessible beyond specialists without dumbing down
- Preserve interpretability as models grow complex—keep reasoning visible, verifiable, questionable
- Design for fluid movement between domains—modeling and making, simulation and synthesis
From OpenEye: modeling extends cognition into invisible domains. Interface shapes what’s thinkable. Design translates between formal systems and intuition.
Acknowledgements
Ashutosh Jogalekar - my visionary product team lead, great friend and mentor. Brent Jenkins, Linda Smyth, Dave Hamilton, Bob Tolbert, Ant Nicholls, Gunther Stahl, Fred Livingston, Amy Migliori, Mirielle Krier, Adam Green, Paul Hawkins, Karen Bridges, Paul George, Bethany Gates, Zeke Ricci, Allison Maresh, Keith Jones, Tim Dunn, John Malloy, Lukas Eberlein, Brenda Montalvo, Sonja Braun Sand, Vincent Vivien, Mark McGann, Chris Bayly… so many other wonderful colleagues, and our excellent customers.
The Bestest Frontend Collaborators Ever: Kevin Schmidt - a friend and mentor I will forever be grateful to, David Estrada, Mireya Rodriguez, Daniel Griego
Video and educational content credit: Vicky Pierce
Docs collaboration: Sarah Roberts, Thom Diaz, David Stevens
Executives: Jeff Grandy, Geoff Skillman, Jharrod LaFonn, Louis Culott
Productive collaboration across frontend/backend leads, TPM, computational chemists. Mutual expertise respect. Shared language. Productive disagreement.
Scientific method and design thinking share structure: both value evidence over opinion, reproducibility, questioning assumptions, learning from failure, iterative improvement.
Technical Environment
Platform: Cloud-native on AWS, NVIDIA NIMS integration, proprietary scheduling for parallel computation
Methods: 3D docking, shape searching, molecular dynamics, free energy calculations, quantum mechanics
Users: Medicinal chemists, computational chemists, drug discovery scientists
Scale: Billions to trillions of molecules
Domain: Pharmaceutical drug discovery, computational chemistry, structure-based design
Databases: Enamine REAL (billions synthesizable), Mcule purchasable compounds, commercial vendors
Performance: FastROCS (1B <30min vs. 11 CPU-years); ROCS X (trillions)